Your new post is loading...
 Your new post is loading...
Researchers have uncovered the intricate molecular mechanism used by parasitic phytoplasma bacteria, known for inducing ‘zombie-like’ effects in plants. This detailed revelation opens new horizons for groundbreaking applications in biotechnology and even in biomedicine. Read full article
This month marks the 10th anniversary of bioRxiv, a significant milestone for the biology preprint server that has revolutionized the way biologists share their research. Here is a personal take on 10 things you should know about preprints. 1. Preprints enable you to share your science at an earlier stage 2. Preprints are citable and should be cited 3. Preprinting is not a movement against peer review 4. Anyone can review preprints 5. Preprints can be be copied and redistributed 6. Preprints are free to publish and free to read 7. Funders should mandate preprints 8. Preprints should not be published when the paper is accepted 9. Preprints should include the supporting data 10. You can preprint anything you want
Open science and transparency serve as antidotes to the epidemic of flawed and fraudulent research. And the journey toward open science begins with preprinting. This post is a celebration of bioRxiv’s 10th anniversary.
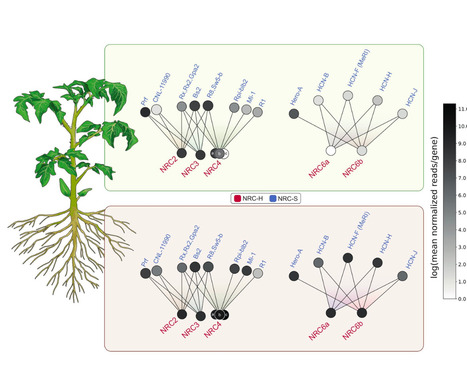
Nucleotide-binding domain and leucine-rich repeat immune receptors (NLRs) confer disease resistance to a multitude of foliar and root parasites of plants. However, the extent to which NLR immunity is expressed differentially between plant organs is poorly known. Here, we show that a large cluster of tomato genes, which encodes the cyst and root-knot nematode disease resistance proteins Hero and MeR1 as well as the NLR-helper NRC6, exhibits nearly exclusive expression in the roots. This root-specific gene cluster emerged in Solanum species about 21 million years ago through gene duplication from the ancient NRC network of asterid plants. NLR-sensors in this gene cluster exclusively signal through NRC6 helpers to trigger the hypersensitive cell death immune response. These findings indicate that the NRC6 gene cluster has sub-functionalized from the larger NRC network to specialize for resistance against root pathogens, including cyst and root-knot nematodes. We propose that NLR gene clusters and networks have evolved organ-specific gene expression as an adaptation to particular parasites and to reduce the risk of autoimmunity.
Via The Sainsbury Lab
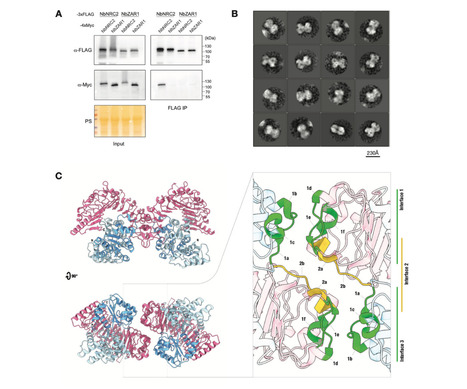
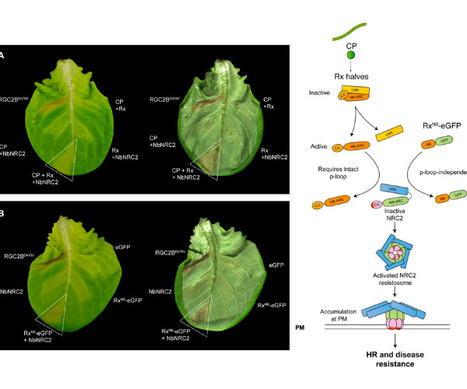
Nucleotide-binding domain and leucine-rich repeat (NLR) proteins can engage in complex interactions to detect pathogens and execute a robust immune response via downstream helper NLRs. However, the biochemical mechanisms of helper NLR activation by upstream sensor NLRs remain poorly understood. Here, we show that the coiled-coil helper NLR NRC2 accumulates in vivo as a homodimer that converts into a higher order oligomer upon activation by 30 its upstream virus disease resistance protein Rx. The Cryo-EM structure of NRC2 in its resting state revealed intermolecular interactions that mediate homodimer formation. These dimerization interfaces have diverged between paralogous NRC proteins to insulate critical network nodes and enable redundant immune pathways. Our results expand the molecular mechanisms of NLR activation pointing to transition from homodimers to higher-order oligomeric resistosomes.
Via The Sainsbury Lab
In the world of science and discovery, luck of course can play a role but it often takes a back seat to dedication, perseverance, and rigorous research. How big is the role of luck in career success? This question has lingered in my mind for decades. I’ve repeatedly been told that I’m lucky — lucky to reside in a Western country, to have secured a job, grants, had a paper accepted, to have exceptional people in my team, to be a good writer and communicator etc. The list goes on, prompting me to reflect — if I tally all these blessings, it seems I must have been touched by a higher power on the day of my birth. The Economist asked the exact same question in a recent column. The article makes a case that career success is significantly influenced by luck, quoting American business magnate and investor Warren Buffett as “winning the ovarian lottery by being born in America”, and “being wired in a way that pays off in a market economy.” Fortunately, the columnists came to their senses and concluded the piece with a more reasonable take:
“If luck can mean a bad decision has a good result, or vice versa, managers should learn to assess the success of an initiative on the basis of process as well as outcome. And if the difference between skill and luck becomes discernible over time, then reward people on consistency of performance, not one-off highs. Mr Buffett might have had a slice of luck at the outset, but a lifetime of investing success suggests he has maximised it.”
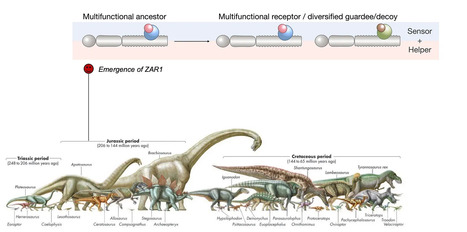
Imagine a plant immune system that has been protecting plants since the time of dinosaurs. Sounds fascinating, right? Well, that’s exactly what we discovered about a plant immune receptor called ZAR1. In our recent study, we delved into the evolutionary history of ZAR1 and uncovered some intriguing findings. ZAR1 is a special type of immune receptor known as an NLR, which helps plants defend against pathogens. What’s remarkable is that while most NLRs evolve rapidly, even within the same species, ZAR1 has been remarkably conserved for millions of years. It traces its origins back to the Jurassic period, around 220 to 150 million years ago, when flowering plants were just starting to emerge.
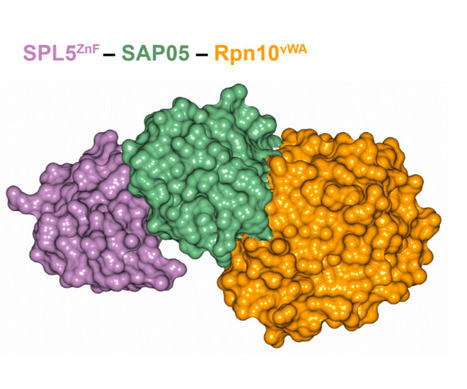
In eukaryotes, targeted protein degradation (TPD) typically depends on a series of interactions among ubiquitin ligases that transfer ubiquitin molecules to substrates leading to degradation by the 26S proteasome. We previously identified that the bacterial effector protein SAP05 mediates ubiquitin-independent TPD. SAP05 forms a ternary complex via interactions with the von Willebrand Factor Type A (vWA) domain of the proteasomal ubiquitin receptor Rpn10 and the zinc-finger (ZnF) domains of the SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) and GATA BINDING FACTOR (GATA) transcription factors (TFs). This leads to direct TPD of the TFs by the 26S proteasome. Here, we report the crystal structures of the SAP05–Rpn10vWA complex at 2.17 Å resolution and of the SAP05–SPL5ZnF complex at 2.20 Å resolution. Structural analyses revealed that SAP05 displays a remarkable bimodular architecture with two distinct nonoverlapping surfaces, a “loop surface” with three protruding loops that form electrostatic interactions with ZnF, and a “sheet surface” featuring two β-sheets, loops, and α-helices that establish polar interactions with vWA. SAP05 binding to ZnF TFs involves single amino acids responsible for multiple contacts, while SAP05 binding to vWA is more stable due to the necessity of multiple mutations to break the interaction. In addition, positioning of the SAP05 complex on the 26S proteasome points to a mechanism of protein degradation. Collectively, our findings demonstrate how a small bacterial bimodular protein can bypass the canonical ubiquitin–proteasome proteolysis pathway, enabling ubiquitin-independent TPD in eukaryotic cells. This knowledge holds significant potential for the creation of TPD technologies.
Via The Sainsbury Lab
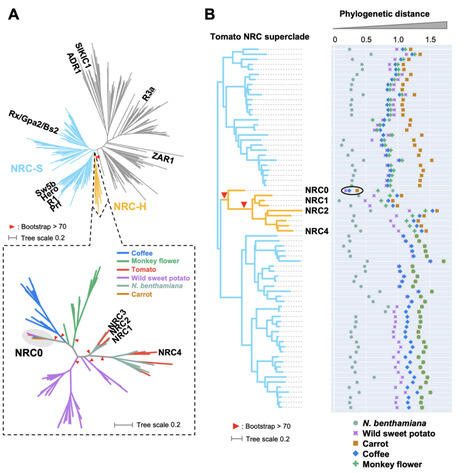
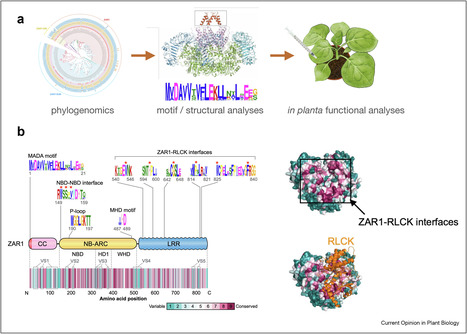
NLR (nucleotide-binding domain and leucine-rich repeat-containing) proteins can form complex receptor networks to confer innate immunity. NRCs are phylogenetically related nodes that function downstream of a massively expanded network of disease resistance proteins that protect against multiple plant pathogens. Here, we used phylogenomic methods to reconstruct the macroevolution of the NRC family. One of the NRCs, we termed NRC0, is the only family member shared across asterid plants, leading us to investigate its evolutionary history and genetic organization. In several asterid species, NRC0 is genetically clustered to other NLRs that are phylogenetically related to NRC-dependent disease resistance genes. This prompted us to hypothesize that the ancestral state of the NRC network is an NLR helper-sensor gene cluster that was present early during asterid evolution. We validated this hypothesis by demonstrating that NRC0 is essential for the hypersensitive cell death induced by its genetically linked sensor NLR partners in four divergent asterid species: tomato, wild sweet potato, coffee and carrot. In addition, activation of a sensor NLR leads to high-order complex formation of its genetically linked NRC0 similar to other NRCs. Our findings map out contrasting evolutionary dynamics in the macroevolution of the NRC network over the last 125 million years from a functionally conserved NLR gene cluster to a massive genetically dispersed network.
Via The Sainsbury Lab
Researchers from Wageningen, Tübingen and Norwich have shed new light on the evolutionary mechanisms that equip wild potato with broad disease resistance against the notorious late blight pathogen (Phytophthora infestans).
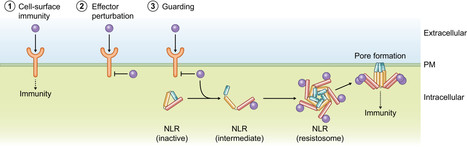
The functional link between cell-surface receptors and intracellular NLR immune receptors is a critical aspect of plant immunity. To establish disease, successful pathogens have evolved mechanisms to suppress cell-surface immune signalling. In response, plants have adapted by evolving NLRs that recognize pathogen effectors involved in this suppression, thereby counteracting their immune-suppressing function. This ongoing co-evolutionary struggle has seemingly resulted in intertwined signalling pathways in some plant species, where NLRs form a separate signalling branch downstream of activated cell-surface receptor complexes essential for full immunity. Understanding these interconnected receptor networks could lead to novel strategies for developing durable disease resistance.
Via The Sainsbury Lab
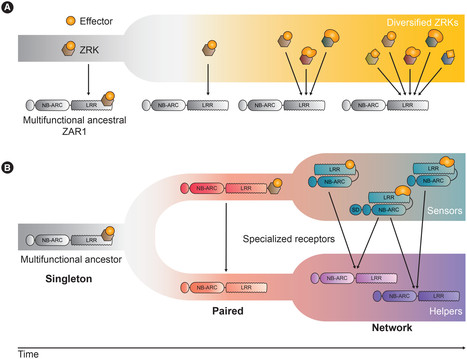
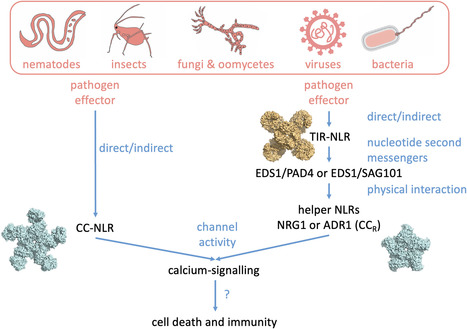
Plants coordinately use cell-surface and intracellular immune receptors to perceive pathogens and mount an immune response. Intracellular events of pathogen recognition are largely mediated by immune receptors of the nucleotide binding and leucine rich-repeat (NLR) classes. Upon pathogen perception, NLRs trigger a potent broad-spectrum immune reaction, usually accompanied by a form of programmed cell death termed the hypersensitive response. Some plant NLRs act as multifunctional singleton receptors which combine pathogen detection and immune signaling. However, NLRs can also function in higher order pairs and networks of functionally specialized interconnected receptors. In this article, we cover the basic aspects of plant NLR biology with an emphasis on NLR networks. We highlight some of the recent advances in NLR structure, function, and activation and discuss emerging topics such as modulator NLRs, pathogen suppression of NLRs, and NLR bioengineering. Multi-disciplinary approaches are required to disentangle how these NLR immune receptor pairs and networks function and evolve. Answering these questions holds the potential to deepen our understanding of the plant immune system and unlock a new era of disease resistance breeding.
Via The Sainsbury Lab
Nucleotide-binding and leucine-rich repeat receptors (NLRs) are a diverse family of intracellular immune receptors that play crucial roles in recognizing and responding to pathogen invasion in plants. This review discusses the overall model of NLR activation and provides an in-depth analysis of the different NLR domains, including N-terminal executioner domains, the nucleotide-binding oligomerization domain (NOD) module, and the leucine-rich repeat (LRR) domain. Understanding the structure-function relationship of these domains is essential for developing effective strategies to improve plant disease resistance and agricultural productivity.
Via The Sainsbury Lab
|
Nicotiana benthamiana is increasingly gaining prominence as a model plant species with recently published high-quality genome assemblies, which will further enable forward and reverse genetic approaches (Bally et al., 2018; Derevnina et al., 2019; Kourelis et al., 2019; Ranawaka et al., 2023; Vollheyde et al., 2023). However, the generation time of N. benthamiana poses a bottleneck in the creation of mutant and transgenic plant lines. Speed breeding (SB), by extended photoperiods and adjustments to growth parameters, is an efficient way to reduce generation times for many crop and model plant species (Ghosh et al., 2018; Watson et al., 2018; Hickey et al., 2019; Varshney et al., 2021). We hypothesized that an extended photoperiod could reduce the seed to seed generation time of N. benthamiana. We tested this hypothesis by comparing generation times under SB conditions to traditionally used photoperiods in growth chambers and green house settings. We found that a 22h photoperiod reduced the generation time of N. benthamiana by approximately 2 weeks (16-22%). Fertilization in combination with a far-red light spectrum did not yield a further reduction in generation time when combined with SB conditions. Our findings further contribute to the establishment of N. benthamiana as an important model organism for plant research.
Via The Sainsbury Lab
Our unwavering obligation, our ‘wajib,’ is to transcend ethnic and national divides and commit ourselves to upholding the fundamental values of human rights and dignity. That’s my dream.
Researchers from The Sainsbury Laboratory and the John Innes Centre in Norwich are excited to contribute to one of the world’s largest crop pathogen surveillance systems as part of an impressive international collaboration constituting 23 institutes. CIMMYT has announced that the initiative is set to expand its analytic and knowledge systems capacity to protect wheat productivity in food vulnerable areas of East Africa and South Asia. Read CIMMYT Press Release
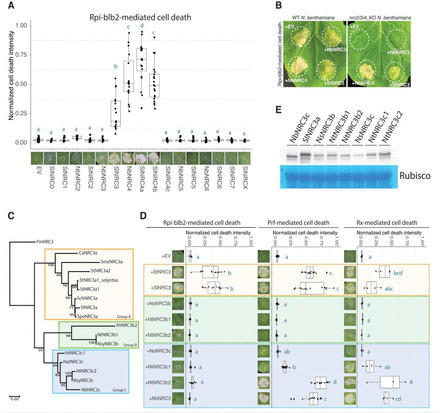
In solanaceous plants, several sensor NLRs and their helper NLRs, known as NRC, form a complex network to confer immunity against pathogens. While the sensor NLRs and downstream NRC helpers display diverse genetic compatibility, the evolution and molecular basis of the complex network structure remained elusive. Here we demonstrated that functional divergence of NRC3 variants has shaped the genetic architecture of the NLR network. Natural NRC3 variants form three allelic groups displaying distinct compatibilities with sensor NLRs. Ancestral sequence reconstruction and analyses of natural and chimeric variants identified six key amino acids involved in sensor-helper compatibility, with two residues critical for subfunctionalization. Co-functioning Rpi-blb2 and NRC3 variants showed stronger transient interactions upon effector detection, with NRC3 membrane-associated complexes forming subsequently. Our findings reveal how mutations in helper NLRs, particularly NRC3, have driven the evolution of their transient interactions with sensor NLRs, leading to subfunctionalization and contributing significantly to the complexity of the NRC network in plant immunity. Teaser Helper NLR subfunctionalization alters transient interactions with sensor NLRs, enhancing plant immune system complexity.
Via The Sainsbury Lab
Nucleotide-binding domain and leucine-rich repeat (NLR) proteins with pathogen sensor activities have evolved to initiate immune signaling by activating helper NLRs. However, the mechanisms underpinning helper NLR activation by sensor NLRs remain poorly understood. Although coiled-coil (CC) type sensor NLRs such as the Potato virus X disease resistance protein Rx have been shown to activate the oligomerization of their downstream helpers NRC2 and NRC4, the domains involved in sensor-helper signaling are not known. Here, we show that the nucleotide binding (NB) domain within the NB-ARC of the Potato virus X disease resistance protein Rx is necessary and sufficient for oligomerization and immune signaling of downstream helper NLRs. In addition, the NB domains of the disease resistance proteins Gpa2 (cyst nematode resistance), Rpi-amr1, Rpi-amr3 (oomycete resistance) and Sw-5b (virus resistance) are also sufficient to activate their respective downstream NRC helpers. Moreover, the NB domain of Rx and its helper NRC2 form a minimal functional unit that can be transferred from solanaceous plants (lamiids) to the Campanulid species lettuce (Lactuca sativa). Our results challenge the prevailing paradigm that NLR proteins exclusively signal via their N-terminal domains and reveal a signaling activity for the NB domain of NRC-dependent sensor NLRs. We propose a model in which helper NLRs monitor the status of the NB domain of their upstream sensors.
Via The Sainsbury Lab
This week, my mind is racing, and I can’t find the zen mood to craft a reflective blog post. Instead, here’s a medley of what’s come across my path. Clickbait silliness In my regular browsing of the Biology topic on AppleNews, I stumbled upon an attention-grabbing headline in BBC Science Focus Magazine — the kind that makes you wonder if you’re about to delve into clickbait silliness. Initially, the article navigates the familiar terrain of speculating how human evolution might unfold should we venture into space. It’s an intriguing notion, albeit based on the somewhat obvious idea that we won’t survive unless we recreate an Earth-like environment (duh!). But then the piece takes a rather bizarre turn, straying into a realm where human biological evolution intersects with speculative notions about societal shifts: “Surrounded by danger and acutely dependent on technology, we might develop much more authoritarian societies where each person must perform their allocated role without question and be ready to sacrifice themselves for the benefit of the species. This would be too important to leave to the unpredictability of democratic, free-market capitalism, so perhaps a rigid hierarchy, akin to the regimes aboard 19th century sailing ships would emerge.” Now, that’s quite the vision. Count me out of that particular interstellar voyage. Yet, sadly aspects of this scenario are unfolding right here on Earth as current events indicate.
Congratulations! You’ve been selected for an in-person interview for a faculty position. Here is 5 tips to help you make a lasting impression during your interview.
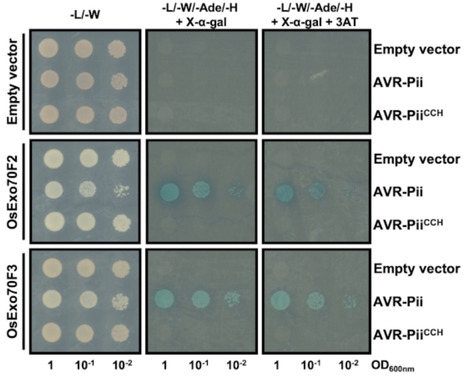
Filamentous plant pathogens deliver effector proteins into host cells to suppress host defence responses and manipulate metabolic processes to support colonization. Understanding the evolution and molecular function of these effectors provides knowledge about pathogenesis and can suggest novel strategies to reduce damage caused by pathogens. However, effector proteins are highly variable, share weak sequence similarity and, although they can be grouped according to their structure, only a few structurally conserved effector families have been functionally characterized to date. Here, we demonstrate that Zinc-finger fold (ZiF) secreted proteins form a functionally diverse effector family in the blast fungus Magnaporthe oryzae. This family relies on the Zinc-finger motif for protein stability and is ubiquitously present, forming different effector tribes in blast fungus lineages infecting 13 different host species. Homologs of the canonical ZiF effector, AVR-Pii from rice infecting isolates, are present in multiple M. oryzae lineages, and the wheat infecting strains of the fungus, for example, possess an allele that also binds host Exo70 proteins and activates the immune receptor Pii. Furthermore, ZiF tribes vary in the host Exo70 proteins they bind, indicating functional diversification and an intricate effector/host interactome. Altogether, we uncovered a new effector family with a common protein fold that has functionally diversified in lineages of M. oryzae. This work expands our understanding of the diversity of M. oryzae effectors, the molecular basis of plant pathogenesis and may ultimately facilitate the development of new sources for pathogen resistance.
Via The Sainsbury Lab
A nurturing academic mentor prioritizes the growth and development of students, but that shouldn’t come at the expense of publishing. And by publishing, I mean all forms of publications.
In the unending coevolutionary dance between plants and microbes, each player impacts the evolution of the other. Here, we provide an overview of the burgeoning field of evolutionary molecular plant–microbe interactions (EVO-MPMI)—the study of mechanisms of plant–microbe interactions in the context of their evolutionary history—tracing its progression from foundational science to practical implementation. We present a snapshot of current research and delve into central concepts, such as conserved features and convergent evolution, as well as methodologies such as ancestral reconstruction. Moreover, we shed light on the practical applications of EVO-MPMI, particularly within the realm of disease control. Looking ahead, we discuss potential future trajectories for EVO-MPMI research, spotlighting the innovative tools and technologies propelling the discipline forward.
Via The Sainsbury Lab
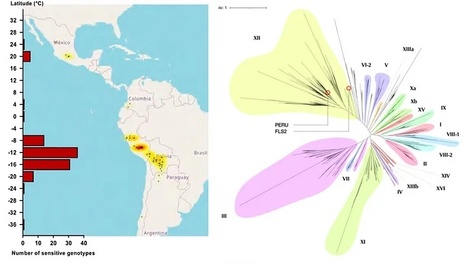
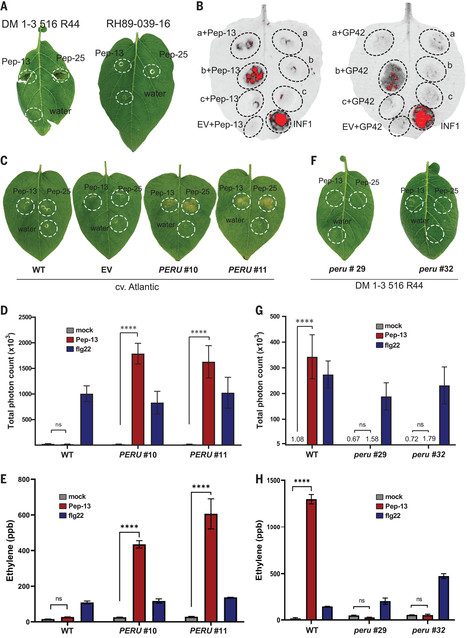
Plant cell surface pattern recognition receptors (PRRs) and intracellular immune receptors cooperate to provide immunity to microbial infection. Both receptor families have coevolved at an accelerated rate, but the evolution and diversification of PRRs is poorly understood. We have isolated potato surface receptor Pep-13 receptor unit (PERU) that senses Pep-13, a conserved immunogenic peptide pattern from plant pathogenic Phytophthora species. PERU, a leucine-rich repeat receptor kinase, is a bona fide PRR that binds Pep-13 and enhances immunity to Phytophthora infestans infection. Diversification in ligand binding specificities of PERU can be traced to sympatric wild tuber-bearing Solanum populations in the Central Andes. Our study reveals the evolution of cell surface immune receptor alleles in wild potato populations that recognize ligand variants not recognized by others.
Via The Sainsbury Lab
The functional link between cell-surface receptors and intracellular NLR immune receptors is a critical aspect of plant immunity. To establish disease, successful pathogens have evolved mechanisms to suppress cell-surface immune signalling. In response, plants have adapted by evolving NLRs that recognize pathogen effectors involved in this suppression, thereby counteracting their immune-suppressing function. This ongoing co-evolutionary struggle has seemingly resulted in intertwined signalling pathways in some plant species, where NLRs form a separate signalling branch downstream of activated cell-surface receptor complexes essential for full immunity. Understanding these interconnected receptor networks could lead to novel strategies for developing durable disease resistance.
Via The Sainsbury Lab
|






 Your new post is loading...
Your new post is loading...