Your new post is loading...
 Your new post is loading...
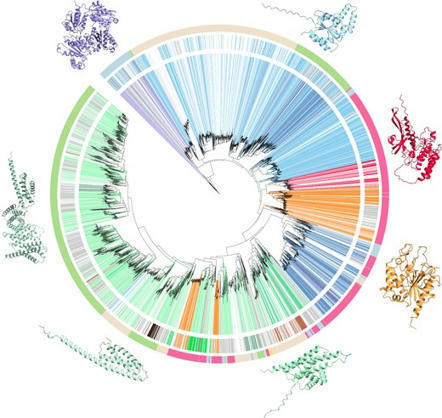
Nucleotide-binding domain and leucine-rich repeat (NLR) proteins are a prominent class of intracellular immune receptors in plants. However, our understanding of plant NLR structure and function is limited to the evolutionarily young flowering plant clade. Here, we describe an extended spectrum of NLR diversity across divergent plant lineages and demonstrate the structural and functional similarities of N-terminal domains that trigger immune responses. We show that the broadly distributed coiled-coil (CC) and toll/interleukin-1 receptor (TIR) domain families of non-flowering plants retain immune-related functions through trans-lineage activation of cell death in the angiosperm Nicotiana benthamiana. We further examined a CC subfamily specific to non-flowering lineages and uncovered an essential N-terminal MAEPL motif that is functionally comparable to motifs in resistosome-forming CC-NLRs. Consistent with a conserved role in immunity, the ectopic activation of CCMAEPL in the non-flowering liverwort Marchantia polymorpha led to profound growth inhibition, defense gene activation, and signatures of cell death. Moreover, comparative transcriptomic analyses of CCMAEPL activity delineated a common CC-mediated immune program shared across evolutionarily divergent non-flowering and flowering plants. Collectively, our findings highlight the ancestral nature of NLR-mediated immunity during plant evolution that dates its origin to at least ∼500 million years ago.
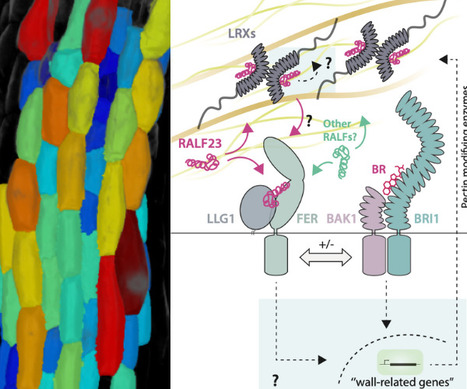
Plant cells survey and modulate their cell wall to control their shape and anisotropic growth. Signaling mediated by the plant steroid hormones brassinosteroids (BR) plays a central role in coordinating cell wall status and cell growth, and alterations in the cell wall BR feedback loop leads to life-threatening defects in tissue and cellular integrity. How the status of the cell wall is relayed to BR signaling remains largely unclear. Increasing evidence shows that RAPID ALKALANIZATION FACTORs (RALFs), a class of secreted peptides, play structural and signaling roles at the cell surface. Here we show that perception of RALF23 promotes the formation and signaling of the main BR receptor complex formed by BRASSINOSTEROID INSENSITIVE 1 (BRI1) and BRI1 BRASSINOSTEROID INSENSITIVE1 BRASSINOSTEROID ASSOCIATED KINASE 1 (BAK1). The loss of the plasma membrane localized RALF receptor complex FERONIA (FER) LORELEI LIKE GPI anchor protein 1 (LLG1) leads to defects in cell expansion and anisotropy, as well as uncontrolled BRI1 BAK1 complex formation and signaling. RALF23 bioactivity relies on pectin status and its perception induces changes in pectin composition and the activity of pectin-modifying enzymes. Our observations suggest a model in which RALF23 functions as a cell wall informed signaling cue initiating a feedback loop that solicits BR signaling, modifies the cell wall, and coordinates cell morphogenesis.
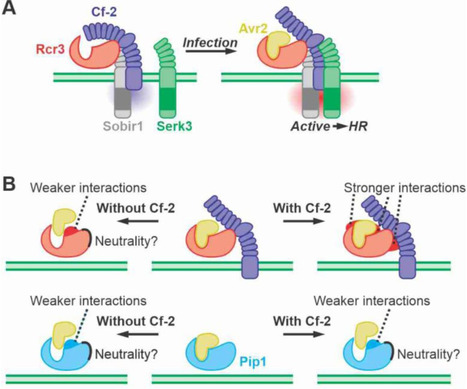
Secreted immune proteases Rcr3 and Pip1 of tomato are both inhibited by Avr2 from the fungal plant pathogen Cladosporium fulvum but only Rcr3 act as a decoy co-receptor that detects Avr2 in the presence of the Cf-2 immune receptor. Here, we identified crucial Rcr3 residues for Cf-2-mediated signalling and bioengineered various proteases to trigger Avr2/Cf-2 dependent immunity. Despite substantial divergences in Rcr3 orthologs from eggplant and tobacco, only minimal alterations were sufficient to trigger Avr2/Cf-2-triggered immune signalling. Tomato Pip1, by contrast, was bioengineered with 16 Rcr3-specific residues to initiate Avr2/Cf-2-triggered immune signalling. These residues cluster on one side next to the substrate binding groove, indicating a potential Cf-2 interaction site. Our findings also revealed that Rcr3 and Pip1 have distinct substrate preferences determined by two variant residues and that both proteases are suboptimal for binding Avr2. This study advances our understanding of the evolution of Avr2 perception and opens avenues to bioengineer proteases to broaden pathogen recognition in other crops.
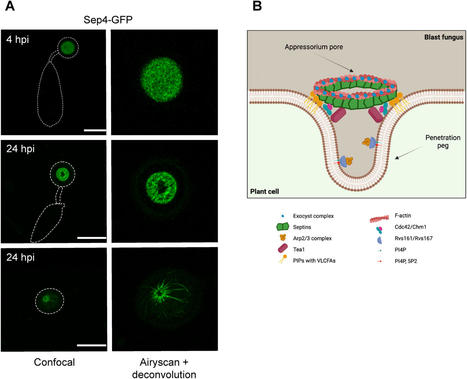
Septin GTPases are morphogenetic proteins that are widely conserved in eukaryotic organisms fulfilling diverse roles in cell division, differentiation and development. In the filamentous fungal pathogen Magnaporthe oryzae, the causal agent of the devastating blast diseases of rice and wheat, septins have been shown to be essential for plant infection. The blast fungus elaborates a specialised infection structure called an appressorium with which it mechanically ruptures the plant cuticle. Septin aggregation and generation of a hetero-oligomeric ring structure at the base of the infection cell is indispensable for plant infection. Furthermore, once the fungus enters host tissue it develops another infection structure, the transpressorium, enabling it to move between living host plant cells, which also requires septins for its function. Specific inhibition of septin aggregation—either genetically or with chemical inhibitors—prevents plant infection. Significantly, by screening for inhibitors of septin aggregation, broad spectrum anti-fungal compounds have been identified that prevent rice blast and a number of other cereal diseases in field trials. We review the recent advances in our understanding of septin biology and their potential as targets for crop disease control.
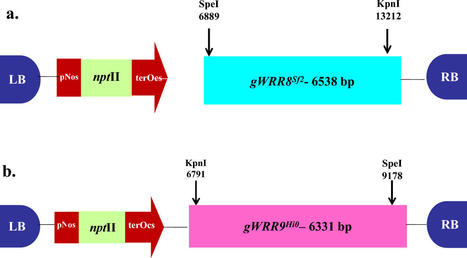
Brassica species is the second most important edible oilseed crop in India. Albugo candida (Pers.) Kuntze, a major oomycete disease of oilseed brassica causing white rust, leads to 60% yield loss globally. The prevalence of A. candida race 2 (Ac2V) that specifically infects B. juncea, coupled with limitations of conventional methods has resulted in a dearth of white rust resistance resources in cultivated varieties. In an effort to develop resistant plants, Agrobacterium mediated genetic transformation of three B. juncea genotypes viz., susceptible host var. Varuna, along with its doubled haploid mutant lines C66 and C69 (showing moderate tolerance to field isolates of A. candida) was initiated to transfer resistance genes (WRR8Sf-2 and WRR9Hi-0) identified in Arabidopsis thaliana against race Ac2V, that encode for Toll-like/interleukin-1 receptor-nucleotide binding-leucine-rich repeat proteins that recognize effectors of the pathogen races. Our results demonstrate that introduction of resistance genes from a tertiary gene pool by genetic transformation enhances disease resistance in B. juncea genotypes to a highly virulent Ac2V isolate.
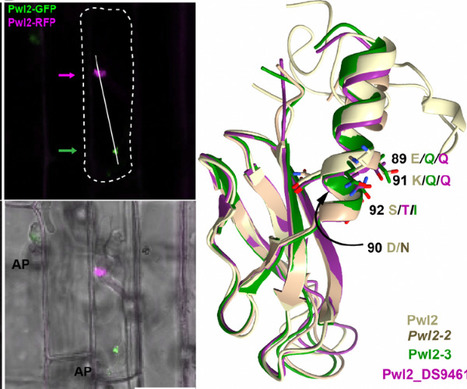
The rice blast fungus Magnaporthe oryzae secretes a battery of effector proteins during host infection to suppress plant immunity and promote disease. Among these effectors, the MAX effector-Pwl2 was first identified as a host specificity determinant for infection of weeping lovegrass (Eragrostis curvula). However, its biological activity has remained unknown. Here we show that the PWL2 gene is ubiquitous in all host-limited forms of M. oryzae and has undergone substantial copy number expansion indicating that it is a core effector of the blast fungus. We used CRISPR/Cas9-mediated gene editing to delete all three copies of PWL2 and generate a pwl2 null mutant in M. oryzae strain Guy11. This resulted in gain-of-virulence towards weeping lovegrass, but a reduction in the severity of disease symptoms on rice. To further investigate the virulence mechanism of Pwl2, we showed that transgenic rice and barley lines constitutively expressing PWL2 display suppression of reactive oxygen species generation and increased susceptibility to infection by M. oryzae. Also, we identify the barley and rice heavy metal-binding isoprenylated protein HIPP43 as a target of the Pwl2 effector. Transgenic lines overexpressing HIPP43 exhibit attenuated defense responses and increased susceptibility to blast infection, consistent with the hypothesis that Pwl2 binds HIPP43 to modulate immunity. This binding is coupled to changes in cellular localisation of these proteins. Taken together, our results provide evidence that Pwl2 is a virulence factor in M. oryzae that acts by suppressing host immunity through binding and re-localising HIPP43.
The availability of sequenced genomes of many agriculturally and commercially important plant species and their disease-causing pathogenic partners is growing. In parallel, there have been several advancements in bioinformatic tools available to predict not only effector proteins (Sperschneider and Dodds 2022) in pathogens but also their targets in host plants from genomic and transcriptomic information (Seong and Krasileva 2023). This wealth of information and tools is leading to the rapid discovery of new effector proteins (Homma et al. 2023; Zhang et al. 2023). However, it is extremely important to experimentally test these predictions both in vitro and in vivo to get meaningful outcomes. Experimental validation will in turn help in the development of better prediction tools.
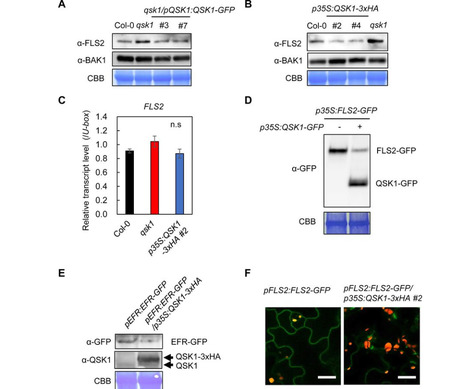
Plants detect pathogens using cell-surface pattern recognition receptors (PRRs) like EFR and FLS2, which recognize bacterial EF-Tu and flagellin, respectively. These PRRs, belonging to the leucine-rich repeat receptor kinase (LRR-RK) family, activate the production of reactive oxygen species via the NADPH oxidase RBOHD. The PRR-RBOHD complex is tightly regulated to prevent unwarranted or exaggerated immune responses. However, certain pathogenic effectors can subvert these regulatory mechanisms, thereby suppressing plant immunity. To elucidate the intricate dynamics of the PRR-RBOHD complex, we conducted a comparative co-immunoprecipitation analysis using EFR, FLS2, and RBOHD. We identified QSK1, an LRR-RK, as a novel component of the PRR-RBOHD complex. QSK1 functions as a negative regulator of PRR-triggered immunity (PTI) by downregulating the abundance of FLS2 and EFR. QSK1 is targeted by the bacterial effector HopF2Pto, a mono-ADP ribosyltransferase, resulting in the reduction of FLS2 and EFR levels through both transcriptional and transcription-independent pathways, thereby inhibiting PTI. Furthermore, HopF2Pto reduces transcript levels of PROSCOOP genes encoding important stress-regulated phytocytokines and their receptor MIK2. Importantly, HopF2Pto requires QSK1 for its accumulation and virulence functions within plants. In summary, our results provide novel insights into the mechanism by which HopF2Pto employs QSK1 to desensitize plants to pathogen attack.
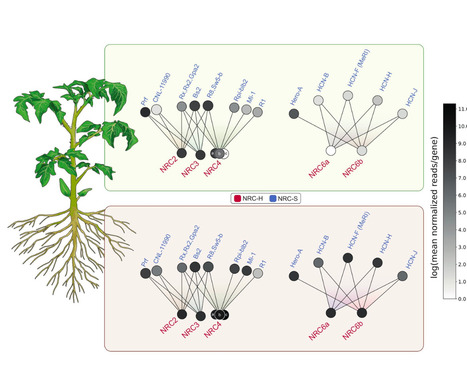
Nucleotide-binding domain and leucine-rich repeat immune receptors (NLRs) confer disease resistance to a multitude of foliar and root parasites of plants. However, the extent to which NLR immunity is expressed differentially between plant organs is poorly known. Here, we show that a large cluster of tomato genes, which encodes the cyst and root-knot nematode disease resistance proteins Hero and MeR1 as well as the NLR-helper NRC6, exhibits nearly exclusive expression in the roots. This root-specific gene cluster emerged in Solanum species about 21 million years ago through gene duplication from the ancient NRC network of asterid plants. NLR-sensors in this gene cluster exclusively signal through NRC6 helpers to trigger the hypersensitive cell death immune response. These findings indicate that the NRC6 gene cluster has sub-functionalized from the larger NRC network to specialize for resistance against root pathogens, including cyst and root-knot nematodes. We propose that NLR gene clusters and networks have evolved organ-specific gene expression as an adaptation to particular parasites and to reduce the risk of autoimmunity.
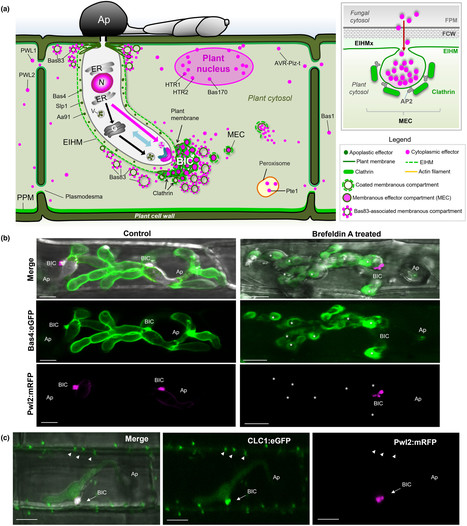
Rice blast, the most destructive disease of cultivated rice world-wide, is caused by the filamentous fungus Magnaporthe oryzae. To cause disease in plants, M. oryzae secretes a diverse range of effector proteins to suppress plant defense responses, modulate cellular processes, and support pathogen growth. Some effectors can be secreted by appressoria even before host penetration, while others accumulate in the apoplast, or enter living plant cells where they target specific plant subcellular compartments. During plant infection, the blast fungus induces the formation of a specialized plant structure known as the biotrophic interfacial complex (BIC), which appears to be crucial for effector delivery into plant cells. Here, we review recent advances in the cell biology of M. oryzae–host interactions and show how new breakthroughs in disease control have stemmed from an increased understanding of effector proteins of M. oryzae are deployed and delivered into plant cells to enable pathogen invasion and host susceptibility.
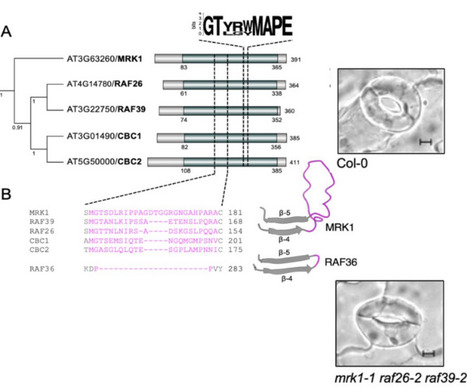
The calcium-dependent protein kinase CPK28 is a regulator of immune homeostasis in multiple plant species. Here, we used a proteomics approach to uncover CPK28-associated proteins. We found that CPK28 associates with subfamily C7 Raf-like kinases MRK1, RAF26, and RAF39, and trans-phosphorylates RAF26 and RAF39. Metazoan Raf kinases function in mitogen-activated protein kinase (MAPK) cascades as MAPK kinase kinases (MKKKs). Although Raf-like kinases share some features with MKKKs, we found that MRK1, RAF26, and RAF39 are unable to trans-phosphorylate any of the 10 Arabidopsis MKKs. We show that MRK1, RAF26, and RAF39 localize to the cytosol and endomembranes, and we define redundant roles for these kinases in stomatal opening, immune-triggered reactive oxygen species (ROS) production, and resistance to a bacterial pathogen. Overall, our study suggests that C7 Raf-like kinases associate with and are phosphorylated by CPK28, function redundantly in stomatal immunity, and possess substrate specificities distinct from canonical MKKKs.
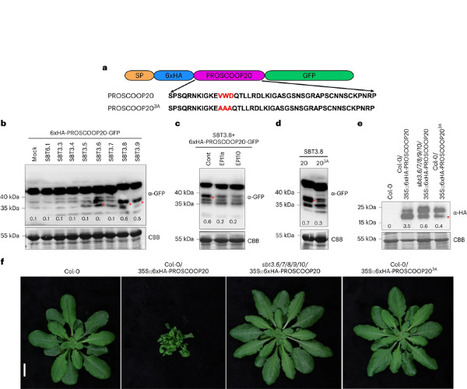
Plant signalling peptides are typically released from larger precursors by proteolytic cleavage to regulate plant growth, development and stress responses. Recent studies reported the characterization of a divergent family of Brassicaceae-specific peptides, SERINE RICH ENDOGENOUS PEPTIDES (SCOOPs), and their perception by the leucine-rich repeat receptor kinase MALE DISCOVERER 1-INTERACTING RECEPTOR-LIKE KINASE 2 (MIK2). Here, we reveal that the SCOOP family is highly expanded, containing at least 50 members in the Columbia-0 reference Arabidopsis thaliana genome. Notably, perception of these peptides is strictly MIK2-dependent. How bioactive SCOOP peptides are produced, and to what extent their perception is responsible for the multiple physiological roles associated with MIK2 are currently unclear. Using N-terminomics, we validate the N-terminal cleavage site of representative PROSCOOPs. The cleavage sites are determined by conserved motifs upstream of the minimal SCOOP bioactive epitope. We identified subtilases necessary and sufficient to process PROSCOOP peptides at conserved cleavage motifs. Mutation of these subtilases, or their recognition motifs, suppressed PROSCOOP cleavage and associated overexpression phenotypes. Furthermore, we show that higher-order mutants of these subtilases show phenotypes reminiscent of mik2 null mutant plants, consistent with impaired PROSCOOP biogenesis, and demonstrating biological relevance of SCOOP perception by MIK2. Together, this work provides insights into the molecular mechanisms underlying the functions of the recently identified SCOOP peptides and their receptor MIK2. The SCOOP signalling peptide family expands to 50 members, whose activities are strictly dependent upon the receptor kinase MIK2. Two subtilase classes process PROSCOOPs, generating bioactive SCOOP peptides. A subtilase mutant phenocopies the mik2 receptor mutant.
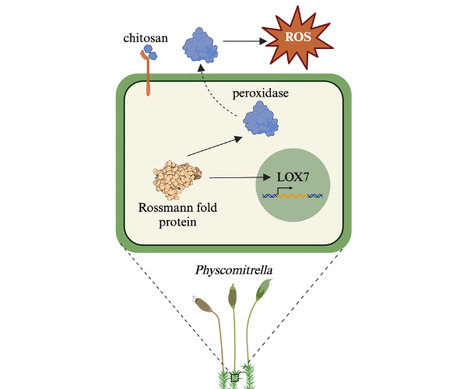
"Despite our current knowledge of early defense responses, more components need to be identified to untangle the signaling cascades leading to activation of PTI in mosses and liverworts. Through the identification and characterization of these key components, a holistic view of plant defense pathways will be obtained. This will facilitate future attempts for engineering more resistant plants. In this issue of Molecular Plant-Microbe Interactions, Marttinen et al. (2023) identified a key gene from Physcomitrella (new botanical name: Physcomitrium patens) in chitosan-induced early immune response."
|
Crop production often faces challenges from plant diseases, and biological control emerges as an effective, environmentally friendly, cost-effective, and sustainable alternative to chemical control. Wheat blast disease caused by fungal pathogen Magnaporthe oryzae Triticum (MoT), is a potential catastrophic threat to global food security. This study aimed to identify potential bacterial isolates from rice and wheat seeds with inhibitory effects against MoT. In dual culture and seedling assays, three bacterial isolates (BTS-3, BTS-4, and BTLK6A) demonstrated effective suppression of MoT growth and reduced wheat blast severity when artificially inoculated at the seedling stage. Genome phylogeny identified these isolates as Bacillus subtilis (BTS-3) and B. velezensis (BTS-4 and BTLK6A). Whole-genome analysis revealed the presence of genes responsible for controlling MoT through antimicrobial defense, antioxidant defense, cell wall degradation, and induced systemic resistance (ISR). Taken together, our results suggest that the suppression of wheat blast disease by seed endophytic B. subtilis (BTS-3) and B. velezensis (BTS-4 and BTLK6A) is liked with antibiosis and induced systemic resistance to wheat plants. A further field validation is needed before recommending these endophytic bacteria for biological control of wheat blast.
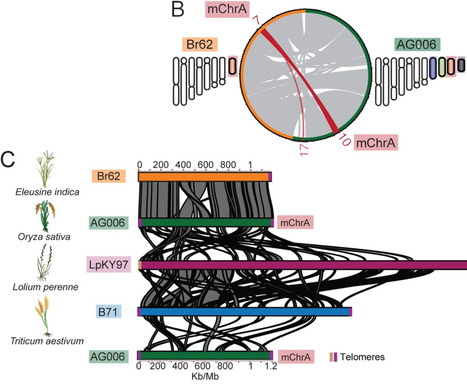
Crop disease pandemics are often driven by clonal lineages of plant pathogens that reproduce asexually. How these clonal pathogens continuously adapt to their hosts despite harboring limited genetic variation, and in absence of sexual recombination remains elusive. Here, we reveal multiple instances of horizontal chromosome transfer within pandemic clonal lineages of the blast fungus Magnaporthe (Syn. Pyricularia) oryzae. We identified a horizontally transferred 1.2Mb supernumerary mini-chromosome which is remarkably conserved between M. oryzae isolates from both the rice blast fungus lineage and the lineage infecting Indian goosegrass (Eleusine indica), a wild grass that often grows in the proximity of cultivated cereal crops. Furthermore, we show that this mini-chromosome was horizontally acquired by clonal rice blast isolates through at least nine distinct transfer events over the past three centuries. These findings establish horizontal mini-chromosome transfer as a mechanism facilitating genetic exchange among different host-associated blast fungus lineages. We propose that blast fungus populations infecting wild grasses act as genetic reservoirs that drive genome evolution of pandemic clonal lineages that afflict cereal crops.
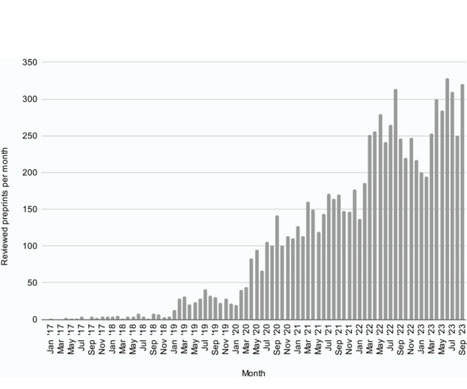
As preprints become more widely used and accepted, they raise the possibility of rethinking the peer-review process. This Consensus View issues a call to action to accelerate the growing momentum of preprint sharing and provides recommendations to empower researchers to provide open and constructive peer review for preprints.
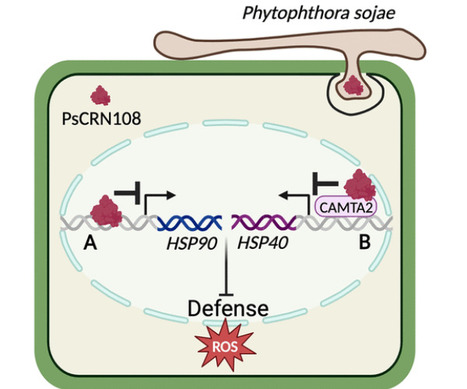
COMMENTARY: The oomycete pathogen, Phytophthora, secretes both apoplastic and cytoplasmic effectors into its plant host upon infection. CRNs (crinkling and necrosis proteins) are a major family of effector proteins characterized by their variable C-terminal domains and conserved N-terminal motifs (Schornack et al. 2010). Although many CRN effectors have been predicted across various oomycete and fungal pathogens, the mechanisms of action of only a few have been characterized.
Nicotiana benthamiana is increasingly gaining prominence as a model plant species with recently published high-quality genome assemblies, which will further enable forward and reverse genetic approaches (Bally et al., 2018; Derevnina et al., 2019; Kourelis et al., 2019; Ranawaka et al., 2023; Vollheyde et al., 2023). However, the generation time of N. benthamiana poses a bottleneck in the creation of mutant and transgenic plant lines. Speed breeding (SB), by extended photoperiods and adjustments to growth parameters, is an efficient way to reduce generation times for many crop and model plant species (Ghosh et al., 2018; Watson et al., 2018; Hickey et al., 2019; Varshney et al., 2021). We hypothesized that an extended photoperiod could reduce the seed to seed generation time of N. benthamiana. We tested this hypothesis by comparing generation times under SB conditions to traditionally used photoperiods in growth chambers and green house settings. We found that a 22h photoperiod reduced the generation time of N. benthamiana by approximately 2 weeks (16-22%). Fertilization in combination with a far-red light spectrum did not yield a further reduction in generation time when combined with SB conditions. Our findings further contribute to the establishment of N. benthamiana as an important model organism for plant research.
Secreted signaling peptides are central regulators of growth, development, and stress responses, but specific steps in the evolution of these peptides and their receptors are not well understood. In addition, the molecular mechanisms of peptide-receptor binding are only known for a few examples, primarily owing to the limited availability of structural capabilities to few laboratories worldwide. Plants have evolved a multitude of secreted signaling peptides and corresponding transmembrane receptors. Stress-responsive SERINE RICH ENDOGENOUS PEPTIDES (SCOOPs) were recently identified. Bioactive SCOOPs are proteolytically processed by subtilases and are perceived by the leucine-rich repeat receptor kinase MALE DISCOVERER 1-INTERACTING RECEPTOR-LIKE KINASE 2 (MIK2) in the model plant Arabidopsis thaliana. How SCOOPs and MIK2 have (co-)evolved, and how SCOOPs bind to MIK2 are however still unknown. Using in silico analysis of 350 plant genomes and subsequent functional testing, we revealed the conservation of MIK2 as SCOOP receptor within the plant order Brassicales. We then leveraged AlphaFold-Multimer and comparative genomics to identify two conserved putative SCOOP-MIK2 binding pockets across Brassicales MIK2 homologues predicted to interact with the ‘SxS’ motif of otherwise sequence-divergent SCOOPs. Notably, mutagenesis of both predicted binding pockets compromised SCOOP binding to MIK2, SCOOP-induced complex formation between MIK2 and its co-receptor BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED KINASE 1 (BAK1), and SCOOP-induced reactive oxygen species production; thus, confirming our in silico predictions. Collectively, in addition to revealing the elusive SCOOP-MIK2 binding mechanisms, our analytic pipeline combining phylogenomics, AI-based structural predictions, and experimental biochemical and physiological validation provides a blueprint for the elucidation of peptide ligand-receptor perception mechanisms.
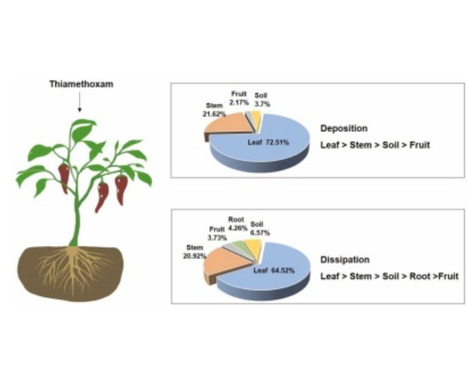
To reduce the application dosage of thiamethoxam (TMX), we investigated the deposition and dissipation patterns in a pepper-planted ecosystem under different planting modes across four regions in China, namely Hainan (HN), Zhejiang (ZJ), Anhui (AH) and Hebei (HB). This study focused on the deposition and dissipation of TMX at concentrations of 63.00, 47.25, 31.50, 23.63 and 15.75 g a.i.hm-2. As the application dose increased, the deposition amount of TMX initially increased in the plants and cultivated soil, showing obvious geographic differences in four cultivation areas. Surprisingly, the initial amount of TMX deposited the pepper-cultivated greenhouse of ZJ and AH was 1.1–2.1-fold and 1.0–3.6-fold higher than that in the open field system at the same application dose, respectively. In pepper leaves, stems, fruits and soil, the dissipation exhibited rapid growth and then slowed. However, the residual concentration showed an increasing trend, followed by a subsequent decrease in the pepper roots. In different planting regions, the dissipation rate of TMX followed the order HN > ZJ > AH > HB in pepper plants and cultivated soil. In comparison to the open field, the total TMX retention rate in greenhouse was higher, indicating overall greater persistence in the greenhouse conditions. These findings reveal the deposition and dissipation characteristics of TMX within the pepper-field ecosystem, offering a significant contribution to the risk assessment of pesticides.
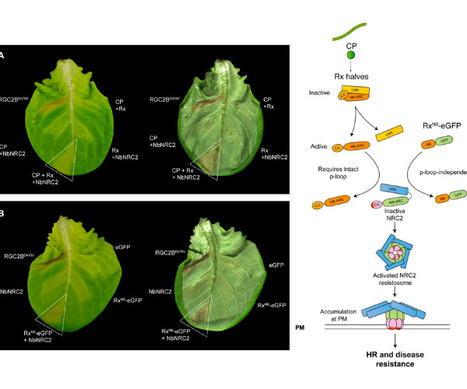
Nucleotide-binding domain and leucine-rich repeat (NLR) proteins can engage in complex interactions to detect pathogens and execute a robust immune response via downstream helper NLRs. However, the biochemical mechanisms of helper NLR activation by upstream sensor NLRs remain poorly understood. Here, we show that the coiled-coil helper NLR NRC2 accumulates in vivo as a homodimer that converts into a higher order oligomer upon activation by 30 its upstream virus disease resistance protein Rx. The Cryo-EM structure of NRC2 in its resting state revealed intermolecular interactions that mediate homodimer formation. These dimerization interfaces have diverged between paralogous NRC proteins to insulate critical network nodes and enable redundant immune pathways. Our results expand the molecular mechanisms of NLR activation pointing to transition from homodimers to higher-order oligomeric resistosomes.
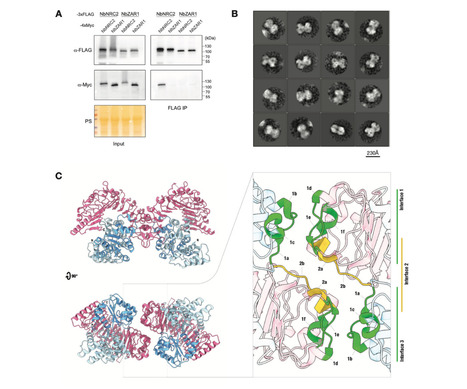
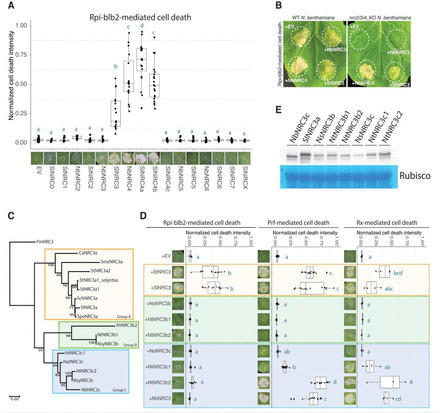
In solanaceous plants, several sensor NLRs and their helper NLRs, known as NRC, form a complex network to confer immunity against pathogens. While the sensor NLRs and downstream NRC helpers display diverse genetic compatibility, the evolution and molecular basis of the complex network structure remained elusive. Here we demonstrated that functional divergence of NRC3 variants has shaped the genetic architecture of the NLR network. Natural NRC3 variants form three allelic groups displaying distinct compatibilities with sensor NLRs. Ancestral sequence reconstruction and analyses of natural and chimeric variants identified six key amino acids involved in sensor-helper compatibility, with two residues critical for subfunctionalization. Co-functioning Rpi-blb2 and NRC3 variants showed stronger transient interactions upon effector detection, with NRC3 membrane-associated complexes forming subsequently. Our findings reveal how mutations in helper NLRs, particularly NRC3, have driven the evolution of their transient interactions with sensor NLRs, leading to subfunctionalization and contributing significantly to the complexity of the NRC network in plant immunity. Teaser Helper NLR subfunctionalization alters transient interactions with sensor NLRs, enhancing plant immune system complexity.
-
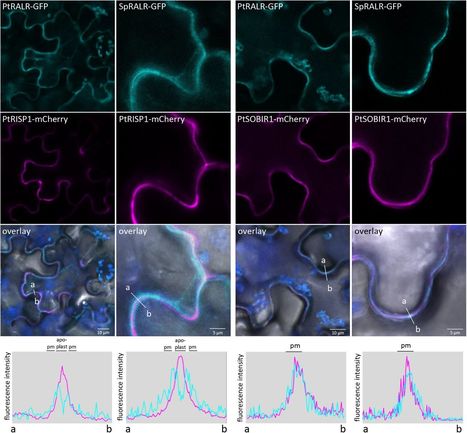
Phytocytokines regulate plant immunity via cell-surface receptors. Populus trichocarpa RUST INDUCED SECRETED PEPTIDE 1 (PtRISP1) exhibits an elicitor activity in poplar, as well as a direct antimicrobial activity against rust fungi. PtRISP1 gene directly clusters with a gene encoding a leucine-rich repeat receptor protein (LRR-RP), that we termed RISP- ASSOCIATED LRR-RP (PtRALR). -
In this study, we used phylogenomics to characterize the RISP and RALR gene families, and functional assays to characterize RISP/RALR pairs. -
Both RISP and RALR gene families specifically evolved in Salicaceae species (poplar and willow), and systematically cluster in the genomes. Two divergent RISPs, PtRISP1 and Salix purpurea RISP1 (SpRISP1), induced a reactive oxygen species (ROS) burst and mitogen- activated protein kinases (MAPKs) phosphorylation in Nicotiana benthamiana leaves expressing the respective clustered RALR. PtRISP1 triggers a rapid stomatal closure in poplar, and both PtRISP1 and SpRISP1 directly inhibit rust pathogen growth. -
Altogether, these results suggest that plants evolved phytocytokines with direct antimicrobial activities, and that the genes coding these phytocytokines co-evolved and physically cluster with their cognate receptors.
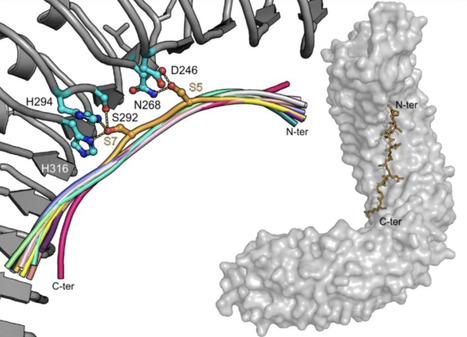
Nucleotide-binding domain and leucine-rich repeat (NLR) proteins with pathogen sensor activities have evolved to initiate immune signaling by activating helper NLRs. However, the mechanisms underpinning helper NLR activation by sensor NLRs remain poorly understood. Although coiled-coil (CC) type sensor NLRs such as the Potato virus X disease resistance protein Rx have been shown to activate the oligomerization of their downstream helpers NRC2 and NRC4, the domains involved in sensor-helper signaling are not known. Here, we show that the nucleotide binding (NB) domain within the NB-ARC of the Potato virus X disease resistance protein Rx is necessary and sufficient for oligomerization and immune signaling of downstream helper NLRs. In addition, the NB domains of the disease resistance proteins Gpa2 (cyst nematode resistance), Rpi-amr1, Rpi-amr3 (oomycete resistance) and Sw-5b (virus resistance) are also sufficient to activate their respective downstream NRC helpers. Moreover, the NB domain of Rx and its helper NRC2 form a minimal functional unit that can be transferred from solanaceous plants (lamiids) to the Campanulid species lettuce (Lactuca sativa). Our results challenge the prevailing paradigm that NLR proteins exclusively signal via their N-terminal domains and reveal a signaling activity for the NB domain of NRC-dependent sensor NLRs. We propose a model in which helper NLRs monitor the status of the NB domain of their upstream sensors.
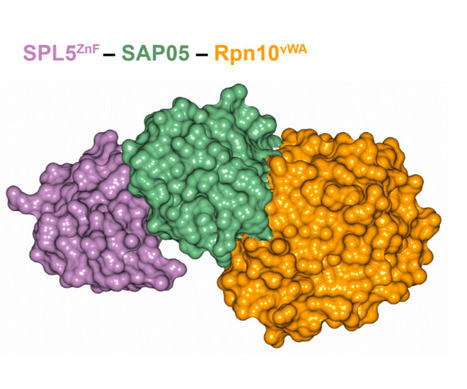
In eukaryotes, targeted protein degradation (TPD) typically depends on a series of interactions among ubiquitin ligases that transfer ubiquitin molecules to substrates leading to degradation by the 26S proteasome. We previously identified that the bacterial effector protein SAP05 mediates ubiquitin-independent TPD. SAP05 forms a ternary complex via interactions with the von Willebrand Factor Type A (vWA) domain of the proteasomal ubiquitin receptor Rpn10 and the zinc-finger (ZnF) domains of the SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) and GATA BINDING FACTOR (GATA) transcription factors (TFs). This leads to direct TPD of the TFs by the 26S proteasome. Here, we report the crystal structures of the SAP05–Rpn10vWA complex at 2.17 Å resolution and of the SAP05–SPL5ZnF complex at 2.20 Å resolution. Structural analyses revealed that SAP05 displays a remarkable bimodular architecture with two distinct nonoverlapping surfaces, a “loop surface” with three protruding loops that form electrostatic interactions with ZnF, and a “sheet surface” featuring two β-sheets, loops, and α-helices that establish polar interactions with vWA. SAP05 binding to ZnF TFs involves single amino acids responsible for multiple contacts, while SAP05 binding to vWA is more stable due to the necessity of multiple mutations to break the interaction. In addition, positioning of the SAP05 complex on the 26S proteasome points to a mechanism of protein degradation. Collectively, our findings demonstrate how a small bacterial bimodular protein can bypass the canonical ubiquitin–proteasome proteolysis pathway, enabling ubiquitin-independent TPD in eukaryotic cells. This knowledge holds significant potential for the creation of TPD technologies.
|





 Your new post is loading...
Your new post is loading...